Archives
- 2018-07
- 2018-10
- 2018-11
- 2019-04
- 2019-05
- 2019-06
- 2019-07
- 2019-08
- 2019-09
- 2019-10
- 2019-11
- 2019-12
- 2020-01
- 2020-02
- 2020-03
- 2020-04
- 2020-05
- 2020-06
- 2020-07
- 2020-08
- 2020-09
- 2020-10
- 2020-11
- 2020-12
- 2021-01
- 2021-02
- 2021-03
- 2021-04
- 2021-05
- 2021-06
- 2021-07
- 2021-08
- 2021-09
- 2021-10
- 2021-11
- 2021-12
- 2022-01
- 2022-02
- 2022-03
- 2022-04
- 2022-05
- 2022-06
- 2022-07
- 2022-08
- 2022-09
- 2022-10
- 2022-11
- 2022-12
- 2023-01
- 2023-02
- 2023-03
- 2023-04
- 2023-05
- 2023-06
- 2023-08
- 2023-09
- 2023-10
- 2023-11
- 2023-12
- 2024-01
- 2024-02
- 2024-03
- 2024-04
- 2024-05
- 2024-06
- 2024-07
- 2024-08
- 2024-09
- 2024-10
- 2024-11
- 2024-12
- 2025-01
- 2025-02
- 2025-03
-
Expression of EBI and its function for migration in
2019-12-05
Expression of EBI2 and its function for migration in vitro in T cells was recently reported (Chalmin et al., 2015, Hannedouche et al., 2011, Liu et al., 2011, Pereira et al., 2009). Pereira et al. (2009) used an EBI2 reporter mouse and found that most CD4+ T cells, but only approximately half of the
-
br Materials and methods Details on
2019-12-05
Materials and methods Details on materials and methods can be found in the Supplemental material section. Results Discussion In our experiments we mimicked inflammation by LPS challenge of M0 macrophages. We discovered a strong increase in expression of CH25H and CYP7B1 while CYP27A1 and H
-
br Acknowledgements The study of RING type
2019-12-04
Acknowledgements The study of RING-type E3s continues to grow extremely rapidly. We regret that it was possible to only cite a fraction of the outstanding primary publications in this field. This work was supported by the National Institute of General Medical Sciences grants R01 GM088055 and R01
-
Koumine HOIP s ability to build linear Ub
2019-12-04
HOIP\'s ability to build linear Ub chains arises from a unique domain that follows directly after the RING2 domain, the linear ubiquitin chain-determining domain (LDD) (Fig. 1). Structures have revealed that the LDD is structurally integrated into RING2, creating a single RING2–LDD unit (Fig. 2D) [4
-
The HEV vaccine developed based on p particles is
2019-12-04

The HEV vaccine, developed based on p239 particles, is of good efficacy, immunogenicity and safety, and was licensed in China in 2012. p239 (aa 368–606) and E2 (aa 394–606) share a common region of ORF2, referred to as E2s (aa 459–606), which harbors the major antigenic determinants of the HEV vacci
-
78 9 In the present work we
2019-12-04
In the present work, we have investigated the reactivity of N-aryl-N′-hydroxyguanidines 1a–d (see Scheme 1 for structures) with the water-soluble Cu(II) complex 8. Using EPR and UV–Visible spectroscopy, we have shown that the studied N-aryl-N′-hydroxyguanidines 1a–d can bind and transfer electrons t
-
Enzymatic assay The usual in vitro test for the
2019-12-04
Enzymatic assay. The usual in vitro test for the measure of the activity of DbH involves ascorbate as a cosubstrate (e.g., 5mM) and tyramine as a substrate (e.g., 10mM). When using N-aryl-N′-hydroxyguanidines instead of ascorbate, the hydroxylase activity of DbH was measured by HPLC as the amounts o
-
S/GSK1349572 br Results br Discussion In this study we
2019-12-04
Results Discussion In this study, we have determined the pathway by which the 3ʹ end of the nascent leading strand is connected with CMGE after priming, revealing that Pol δ likely plays a crucial role in establishing all continuously synthesized leading strands at eukaryotic replication origi
-
br Conclusions In this paper we describe the recombinant exp
2019-12-04
Conclusions In this paper we describe the recombinant expression and characterization of ParI, a C5-DNA-MTase from P. arcticus 273–4. To our knowledge, this is the first characterization of an orphan C5-DNA-MTase from a psychrophilic bacterium. The C5-DNA-MTase could not be expressed in regular E
-
Ligand independent constitutively active variants have also
2019-12-04
Ligand-independent, constitutively active variants have also been found in receptors with associated tyrosine kinases. In approximately 60% of patients harboring inflammatory hepatocellular adenomas (IHCAs), these adenomas have been caused by small in-frame deletions in the cytokine-binding domain o
-
Four m thick sections of formalin fixed paraffin embedded
2019-12-03
Four-μm thick sections of formalin-fixed paraffin-embedded tissue samples from uterine cervical cancers were cut with a microtome and dried overnight at 37 °C on a silanized-slide (Dako, Carpentaria, CA, USA). The protocol of the universal Dako Labeled Streptavidin–Biotin kit (Dako) was followed for
-
br Experimental br Results and discussion br Conclusion
2019-12-03
Experimental Results and discussion Conclusion Acknowledgements This work was funded by the National Natural Science Foundation of China (81572080, 81873972and81873980), the National Science and Technology Major Project of the Ministry of Science and Technology of China (2018ZX10732202),
-
Phenolic compounds generated during biomass pretreatment inh
2019-12-03
Phenolic compounds generated during TNF-alpha, recombinant murine protein pretreatment inhibit and/or deactivate cellulolytic/hemicellulolytic enzymes as well as the viability and fermentative capacity of yeast and bacteria [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]. Phenolic compounds ca
-
br Materials and methods br Results and discussion br
2019-12-03
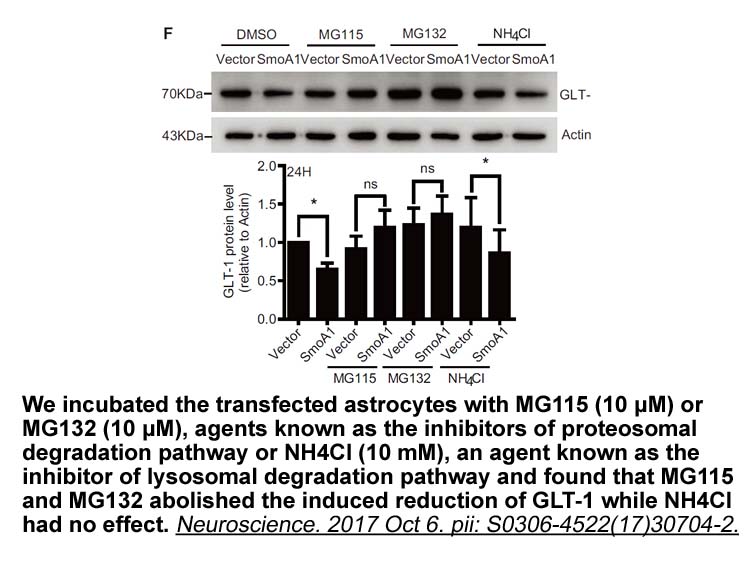
Materials and methods Results and discussion Concluding remarks Enzymatic production of bioactive 9-cis-11-trans-conjugated linoleic Amodiaquine dihydrochloride dihydrate receptor is part of a detoxification mechanism against linoleic acid which is a toxic and stress factor for many lactic
-
mglur In principle in vitro techniques for
2019-12-03
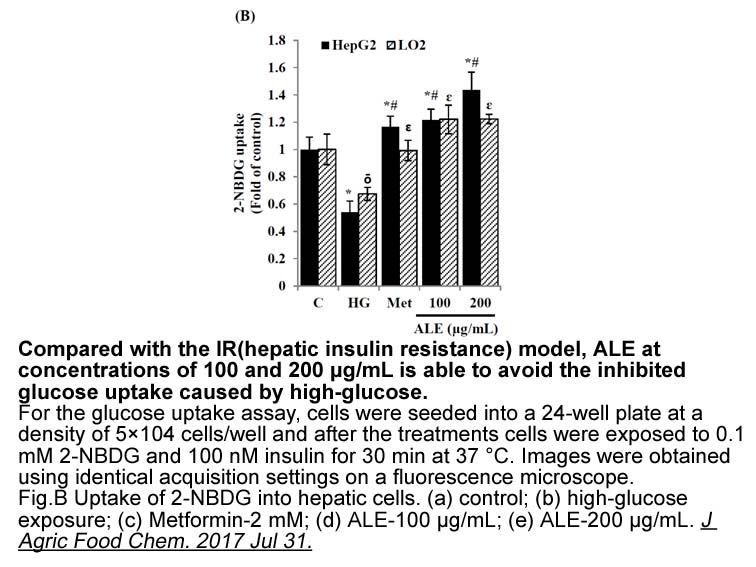
In principle, in vitro techniques for assessing enzyme induction might include the use of cultured hepatocytes, precision-cut liver slices, immortalized mglur lines (such as those derived from hepatomas) and reporter gene constructs (where appropriate cells are transfected with recombinant DNA encod
15753 records 893/1051 page Previous Next First page 上5页 891892893894895 下5页 Last page